Quickstart example
Georg Rüppel
2024-12-04
Source:vignettes/movetrack_example.Rmd
movetrack_example.RmdThis is a quickstart guide to model and visualize animal movements
from radio-telemetry data using the movetrack package.
library(movetrack)
library(ggplot2)
theme_set(theme_bw(base_size = 15))
# Load example data
data(motusData)Estimate locations
The first step involves estimating animal locations based on antenna
bearings and signal strength at two-minute intervals using the
locate function.
loc <- locate(motusData, dTime = 2)## Removed 10 detections containing missing values.Model flight paths
Next, we model the animal’s flight paths using Markov Chain Monte
Carlo (MCMC) with four parallel chains utilising the track
function and return a summary of the results.
fit <- track(loc, parallel_chains = 4, refresh = 1e3)## Running MCMC with 4 parallel chains...
##
## Chain 1 Iteration: 1 / 2000 [ 0%] (Warmup)## Chain 2 Iteration: 1 / 2000 [ 0%] (Warmup)## Chain 3 Iteration: 1 / 2000 [ 0%] (Warmup)## Chain 4 Iteration: 1 / 2000 [ 0%] (Warmup)## Chain 3 Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3 Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1 Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1 Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4 Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4 Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3 Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3 finished in 128.7 seconds.
## Chain 2 Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2 Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1 Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1 finished in 150.3 seconds.
## Chain 2 Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2 finished in 171.0 seconds.
## Chain 4 Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4 finished in 185.8 seconds.
##
## All 4 chains finished successfully.
## Mean chain execution time: 158.9 seconds.
## Total execution time: 185.9 seconds.
fitClick to expand
## ID time lon lat distance speed
## 1 49237 2023-08-20 20:16:00 8.80 54.8 NA NA
## 2 49237 2023-08-20 20:18:00 8.80 54.8 1110 9.25
## 3 49237 2023-08-20 20:20:00 8.79 54.8 1358 11.32
## 4 49237 2023-08-20 20:22:00 8.79 54.8 1476 12.30
## 5 49237 2023-08-20 20:24:00 8.78 54.8 1571 13.09
## 6 49237 2023-08-20 20:26:00 8.77 54.8 1681 14.01
## 7 49237 2023-08-20 20:28:00 8.76 54.8 1876 15.63
## 8 49237 2023-08-20 20:30:00 8.75 54.8 2021 16.84
## 9 49237 2023-08-20 20:32:00 8.74 54.7 1920 16.00
## 10 49237 2023-08-20 20:34:00 8.74 54.7 1804 15.04
## 11 49237 2023-08-20 20:36:00 8.74 54.7 1733 14.44
## 12 49237 2023-08-20 20:38:00 8.75 54.7 1680 14.00
## 13 49237 2023-08-20 20:40:00 8.76 54.7 1567 13.06
## 14 49237 2023-08-20 20:42:00 8.77 54.7 1425 11.87
## 15 49237 2023-08-20 20:44:00 8.78 54.7 1364 11.36
## 16 49237 2023-08-20 20:46:00 8.78 54.7 1375 11.46
## 17 49237 2023-08-20 20:48:00 8.79 54.7 1531 12.75
## 18 49237 2023-08-20 20:50:00 8.80 54.6 1847 15.39
## 19 49237 2023-08-20 20:52:00 8.80 54.6 2191 18.26
## 20 49237 2023-08-20 20:54:00 8.81 54.6 2333 19.44
## 21 49237 2023-08-20 20:56:00 8.81 54.6 2294 19.11
## 22 49237 2023-08-20 20:58:00 8.82 54.6 2444 20.37
## 23 49237 2023-08-20 21:00:00 8.82 54.5 2738 22.82
## 24 49237 2023-08-20 21:08:00 8.83 54.5 2919 6.08
## 25 49237 2023-08-20 21:10:00 8.82 54.5 2701 22.51
## 26 49237 2023-08-20 21:12:00 8.82 54.5 2228 18.57
## 27 49237 2023-08-20 21:14:00 8.81 54.5 1910 15.91
## 28 49237 2023-08-20 21:16:00 8.79 54.4 1770 14.75
## 29 49237 2023-08-20 21:18:00 8.78 54.4 1698 14.15
## 30 49237 2023-08-20 21:20:00 8.77 54.4 1698 14.15
## 31 49237 2023-08-20 21:22:00 8.76 54.4 1811 15.09
## 32 49237 2023-08-20 21:24:00 8.75 54.4 1958 16.32
## 33 49237 2023-08-20 21:26:00 8.75 54.4 1844 15.37
## 34 49237 2023-08-20 21:28:00 8.75 54.4 1708 14.23
## 35 49237 2023-08-20 21:30:00 8.76 54.4 1650 13.75
## 36 49237 2023-08-20 21:32:00 8.76 54.3 1702 14.18
## 37 49237 2023-08-20 21:34:00 8.76 54.3 1717 14.31
## 38 49237 2023-08-20 21:36:00 8.77 54.3 1603 13.36
## 39 49237 2023-08-20 21:38:00 8.77 54.3 1429 11.91
## 40 49237 2023-08-20 21:40:00 8.77 54.3 1404 11.70
## 41 49237 2023-08-20 21:42:00 8.77 54.3 1539 12.83
## 42 49237 2023-08-20 21:44:00 8.77 54.3 1804 15.03
## 43 49237 2023-08-20 21:46:00 8.77 54.3 2142 17.85
## 44 49237 2023-08-20 21:48:00 8.77 54.2 2258 18.82
## 45 49237 2023-08-20 21:50:00 8.77 54.2 2493 20.78
## 46 49237 2023-08-20 21:52:00 8.76 54.2 2812 23.43
## 47 49237 2023-08-20 22:10:00 8.76 54.2 2740 2.54
## 48 49237 2023-08-20 22:12:00 8.75 54.1 2443 20.36
## 49 49237 2023-08-20 22:14:00 8.74 54.1 2054 17.11
## 50 49237 2023-08-20 22:16:00 8.73 54.1 2017 16.81
## 51 49237 2023-08-20 22:24:00 8.72 54.1 2303 4.80
## 52 49237 2023-08-20 22:26:00 8.71 54.1 2695 22.46
## 53 49237 2023-08-20 22:28:00 8.70 54.0 3364 28.03
## 54 49237 2023-08-20 22:30:00 8.69 54.0 4277 35.65
## 55 49237 2023-08-20 22:34:00 8.68 54.0 5311 22.13
## 56 49237 2023-08-20 23:00:00 8.65 53.9 5980 3.83
## 57 49237 2023-08-20 23:02:00 8.63 53.9 6718 55.98
## 58 49237 2023-08-20 23:04:00 8.59 53.8 7558 62.98
## 59 49237 2023-08-21 00:16:00 8.55 53.7 8155 1.89
## 60 49237 2023-08-21 00:18:00 8.50 53.7 8341 69.51
## 61 49237 2023-08-21 00:20:00 8.45 53.6 8066 67.22
## 62 49237 2023-08-21 00:22:00 8.40 53.5 7826 65.22
## 63 49237 2023-08-21 00:24:00 8.34 53.5 7748 64.57
## 64 49237 2023-08-21 00:26:00 8.28 53.4 7966 66.39
## 65 49237 2023-08-21 01:44:00 8.23 53.3 7924 1.69
## 66 49237 2023-08-21 01:46:00 8.18 53.3 7424 61.87
## 67 49237 2023-08-21 01:48:00 8.14 53.2 6464 53.87
## 68 49237 2023-08-21 01:50:00 8.11 53.2 5435 45.30
## 69 49237 2023-08-21 01:52:00 8.08 53.1 4566 38.05
## 70 49237 2023-08-21 01:54:00 8.06 53.1 4083 34.02
## 71 49237 2023-08-21 01:56:00 8.04 53.1 3805 31.71
## 72 50893 2023-10-26 18:38:00 8.78 54.3 NA NA
## 73 50893 2023-10-26 18:40:00 8.78 54.3 4512 37.60
## 74 50893 2023-10-26 18:42:00 8.78 54.2 6349 52.91
## 75 50893 2023-10-26 18:44:00 8.79 54.2 7782 64.85
## 76 50893 2023-10-26 18:46:00 8.80 54.2 4161 34.67
## 77 50893 2023-10-26 18:48:00 8.81 54.2 3563 29.69
## 78 50893 2023-10-26 18:50:00 8.82 54.2 3549 29.58
## 79 50893 2023-10-26 18:52:00 8.84 54.2 3582 29.85
## 80 50893 2023-10-26 18:54:00 8.86 54.2 3786 31.55
## 81 50893 2023-10-26 18:56:00 8.87 54.1 4128 34.40
## 82 50893 2023-10-26 18:58:00 8.88 54.1 4686 39.05
## 83 50893 2023-10-26 19:00:00 8.88 54.1 4778 39.82
## 84 50893 2023-10-26 19:02:00 8.88 54.0 4666 38.88
## 85 50893 2023-10-26 19:04:00 8.86 54.0 4434 36.95
## 86 50893 2023-10-26 19:06:00 8.82 54.0 4008 33.40
## 87 50893 2023-10-26 19:08:00 8.77 54.0 4874 40.62
## 88 50893 2023-10-26 19:10:00 8.67 54.0 7373 61.44
## 89 50893 2023-10-26 19:12:00 8.53 53.9 12532 104.44
## 90 50893 2023-10-26 19:50:00 8.37 53.8 15571 6.83
## 91 50893 2023-10-26 19:52:00 8.24 53.7 13940 116.16
## 92 50893 2023-10-26 19:54:00 8.19 53.7 7902 65.85
## 93 50893 2023-10-26 19:56:00 8.15 53.6 5129 42.74
## 94 50893 2023-10-26 19:58:00 8.10 53.6 4446 37.05
## 95 50893 2023-10-26 20:00:00 8.07 53.6 3563 29.69
## 96 50893 2023-10-26 20:02:00 8.05 53.6 3443 28.69
## 97 50893 2023-10-26 20:04:00 8.05 53.6 3243 27.03
## 98 50893 2023-10-26 20:06:00 8.06 53.6 3279 27.33
## 99 50893 2023-10-26 20:08:00 8.07 53.6 3555 29.62
## 100 50893 2023-10-26 20:10:00 8.07 53.6 3384 28.20
## 101 50893 2023-10-26 20:12:00 8.00 53.7 6202 51.69
## 102 50893 2023-10-26 20:14:00 7.80 53.7 13819 115.16
## 103 50893 2023-10-26 20:16:00 7.75 53.7 5640 47.00
## 104 50893 2023-10-26 20:18:00 7.73 53.7 3564 29.70
## 105 50893 2023-10-26 20:20:00 7.65 53.7 6875 57.29
## 106 50893 2023-10-26 20:34:00 7.44 53.7 14031 16.70
## 107 50893 2023-10-26 20:36:00 7.26 53.7 12533 104.44
## 108 50893 2023-10-26 20:38:00 7.22 53.7 4187 34.89
## 109 50893 2023-10-26 20:40:00 7.22 53.7 3086 25.72
## 110 50893 2023-10-26 20:42:00 7.23 53.7 3014 25.12
## 111 50893 2023-10-26 20:44:00 7.23 53.7 2993 24.94
## 112 50893 2023-10-26 20:46:00 7.22 53.7 3094 25.78
## 113 50893 2023-10-26 20:48:00 7.20 53.6 3333 27.77
## 114 50893 2023-10-26 20:50:00 7.19 53.6 3276 27.30
## 115 50893 2023-10-26 20:52:00 7.16 53.6 3744 31.20
## 116 50893 2023-10-26 20:54:00 7.14 53.6 3947 32.89
## 117 50893 2023-10-26 20:56:00 7.13 53.6 4484 37.37
## 118 50893 2023-10-26 21:04:00 7.09 53.6 5591 11.65
## 119 50893 2023-10-26 21:06:00 7.08 53.5 4463 37.19
## 120 50893 2023-10-26 21:08:00 7.08 53.5 3502 29.19
## 121 50893 2023-10-26 21:10:00 7.08 53.5 3193 26.61
## 122 50893 2023-10-26 21:12:00 7.08 53.5 3124 26.03
## 123 50893 2023-10-26 21:14:00 7.08 53.5 3047 25.39
## 124 50893 2023-10-26 21:20:00 7.08 53.5 3634 10.09
## 125 50893 2023-10-26 21:22:00 7.09 53.5 3403 28.36
## 126 50893 2023-10-26 21:24:00 7.09 53.5 3505 29.21
## 127 50893 2023-10-26 21:26:00 7.08 53.5 5099 42.49
## 128 50893 2023-10-26 21:28:00 6.93 53.4 13121 109.34
## 129 50893 2023-10-26 21:30:00 6.51 53.3 30901 257.50
## 130 50893 2023-10-26 23:26:00 5.92 53.2 41977 6.03
## 131 50893 2023-10-26 23:28:00 5.35 53.1 40417 336.81
## 132 50893 2023-10-26 23:30:00 4.99 53.0 26002 216.68
## 133 50893 2023-10-26 23:38:00 4.84 52.9 12071 25.15
## 134 50893 2023-10-26 23:40:00 4.82 52.9 4532 37.77
## 135 50893 2023-10-26 23:42:00 4.83 52.9 3690 30.75
## 136 50893 2023-10-26 23:44:00 4.82 52.9 3397 28.31
## 137 50893 2023-10-26 23:46:00 4.81 52.9 3565 29.71
## 138 50893 2023-10-26 23:48:00 4.78 52.9 3955 32.96
## 139 50893 2023-10-26 23:50:00 4.76 52.9 3705 30.88
## 140 50893 2023-10-26 23:52:00 4.75 52.9 3658 30.49
## 141 50893 2023-10-26 23:54:00 4.74 52.9 3705 30.87
## 142 50893 2023-10-26 23:56:00 4.71 52.9 3827 31.89
## 143 50893 2023-10-26 23:58:00 4.68 52.8 3736 31.13
## 144 50893 2023-10-27 00:00:00 4.67 52.8 3572 29.77
## 145 50893 2023-10-27 00:02:00 4.67 52.8 3548 29.57
## 146 50893 2023-10-27 00:04:00 4.67 52.8 3565 29.71
## 147 50893 2023-10-27 00:06:00 4.64 52.8 4090 34.08
## 148 50893 2023-10-27 00:08:00 4.62 52.8 4106 34.22
## 149 50893 2023-10-27 00:10:00 4.61 52.8 3730 31.08
## 150 50893 2023-10-27 00:12:00 4.59 52.8 3623 30.19
## 151 50893 2023-10-27 00:14:00 4.59 52.8 3675 30.62
## 152 50893 2023-10-27 00:16:00 4.57 52.7 5006 41.72
## 153 50893 2023-10-27 00:18:00 4.55 52.7 8544 71.20
## 154 50893 2023-10-27 00:46:00 4.53 52.6 7474 4.45
## 155 50893 2023-10-27 00:48:00 4.53 52.6 5539 46.16
## 156 50893 2023-10-27 00:50:00 4.52 52.6 3938 32.82
## 157 50893 2023-10-27 00:52:00 4.52 52.6 3553 29.61
## 158 50893 2023-10-27 00:54:00 4.52 52.6 3610 30.08
## 159 50893 2023-10-27 00:56:00 4.53 52.6 3535 29.46
## 160 50893 2023-10-27 00:58:00 4.53 52.6 3642 30.35
## 161 50893 2023-10-27 01:00:00 4.53 52.6 4498 37.48Plot flight speed
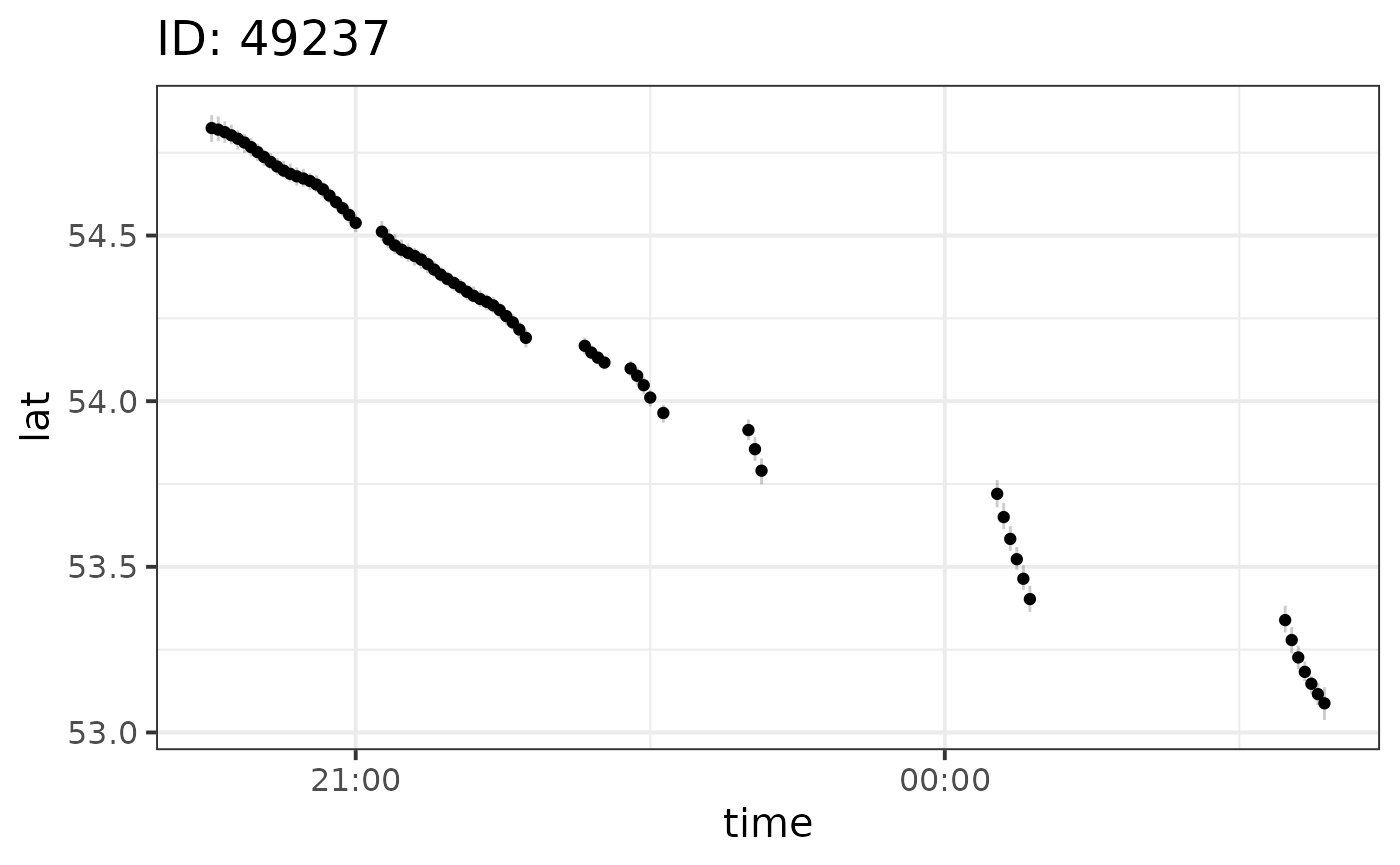
We can plot the results per individual and output variable using the
plot function:
plot(fit, id = 49237)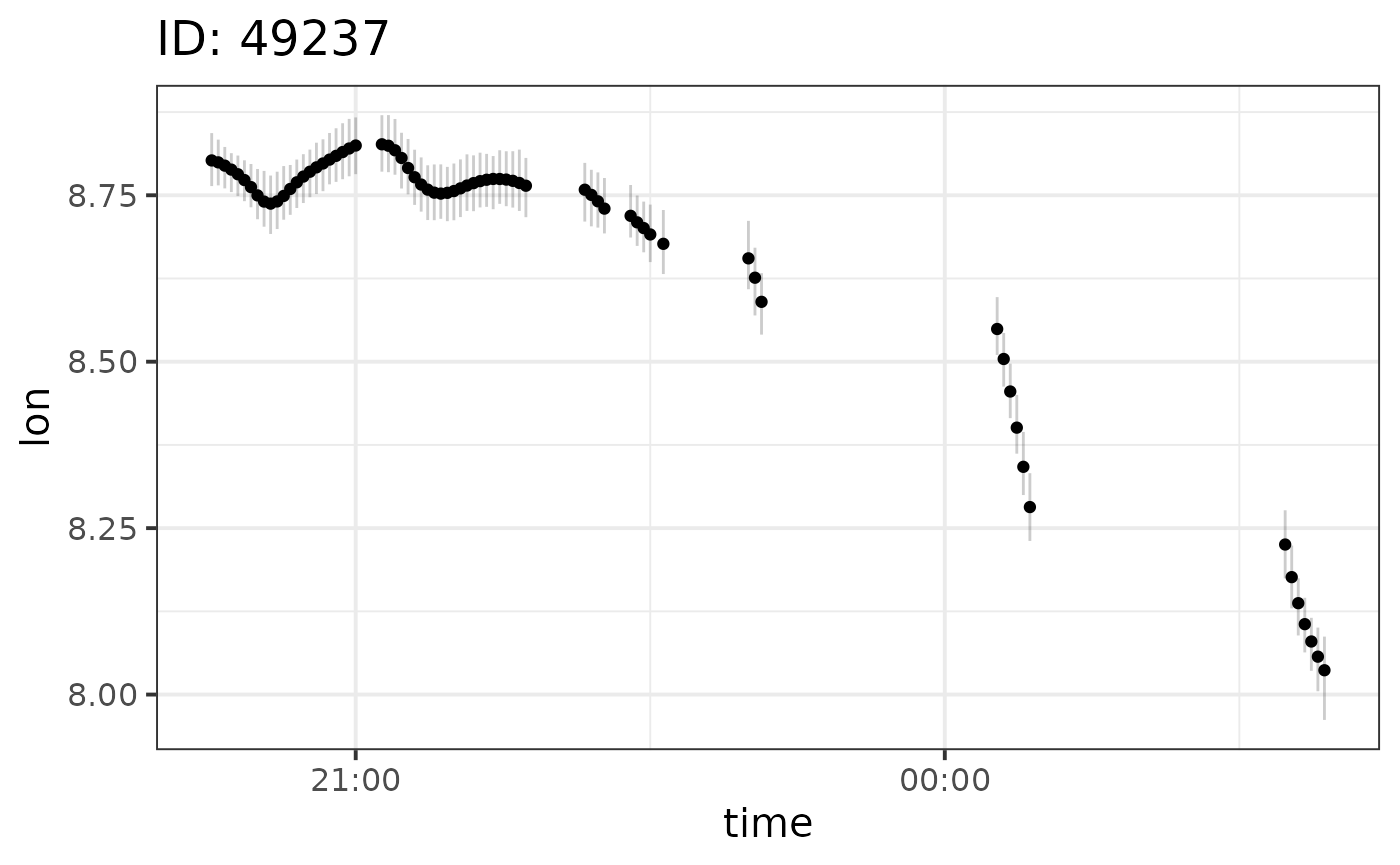
Plot flight paths
We can visualise the results on a map using the mapTrack
function:
mapTrack(fit) +
geom_point(aes(recvDeployLon, recvDeployLat), data = motusData)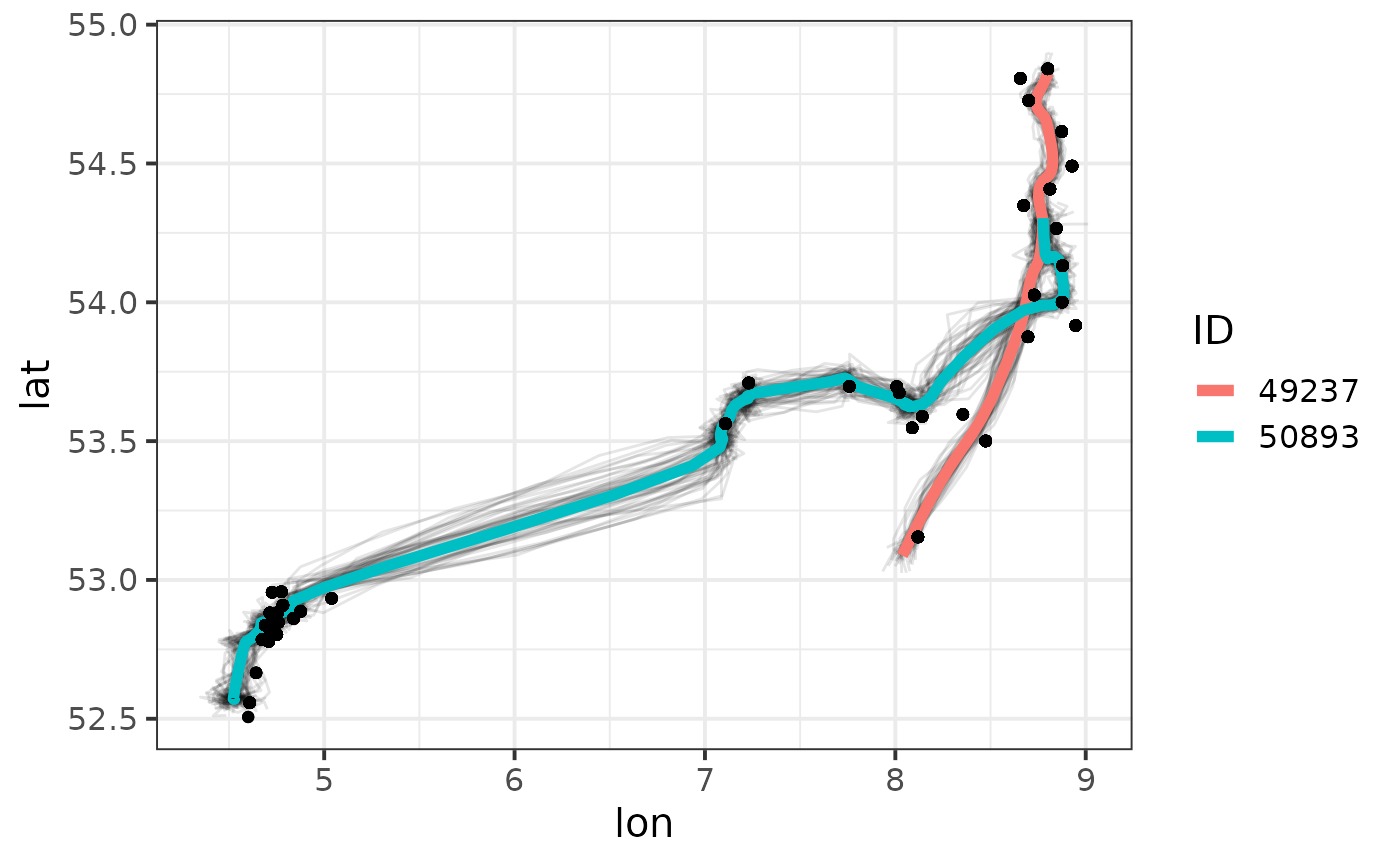
Modelled movement trajectories per individual. Posterior means are shown together with 50 posterior draws, circles on the map indicate receiver locations with detections of the animals.
We could also create a Leaflet map using the following code:
library(sfheaders)
library(leaflet)
# Extract draws
draws <- getDraws(fit) |>
sf_linestring("lon", "lat", linestring_id = "tID")
# Leaflet map
fit |>
as.data.frame() |>
sf_linestring("lon", "lat", linestring_id = "ID") |>
leaflet() |>
addTiles() |>
addPolylines(data = draws, color = "grey", weight = 1, opacity = 0.2) |>
addPolylines(color = ~ c("orange", "blue"))